 A1.1: Simulation and optimization of VIPER uncaging
A1.1: Simulation and optimization of VIPER uncaging
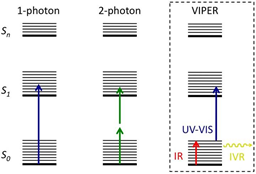 In collaboration with the Bredenbeck group (A4) and the Heckel group (A2), this PhD thesis will systematically contribute to a cycle of theoretical prediction, spectroscopic investigation, and synthesis, in order to optimize novel uncaging strategies. The PhD student of this project will focus upon the Vibrationally Promoted Electronic Resonance (VIPER) experiment pioneered by the Bredenbeck group (A4) that potentially offers an ideal tool for orthogonal uncaging, due to highly selective excitation in the mid-IR range, followed by a UV-Vis pulse.
In collaboration with the Bredenbeck group (A4) and the Heckel group (A2), this PhD thesis will systematically contribute to a cycle of theoretical prediction, spectroscopic investigation, and synthesis, in order to optimize novel uncaging strategies. The PhD student of this project will focus upon the Vibrationally Promoted Electronic Resonance (VIPER) experiment pioneered by the Bredenbeck group (A4) that potentially offers an ideal tool for orthogonal uncaging, due to highly selective excitation in the mid-IR range, followed by a UV-Vis pulse.
On the theory side, the PhD student will establish a simulation protocol for the VIPER experiment in order to assist in its interpretation and optimisation, as well as the prediction of optimal orthogonal cages. Several steps are involved in the analysis, including (i) the calculation of vibronic spectra as a function of vibrational excitation in the electronic ground state, (ii) the analysis of the influence of intramolecular vibrational redistribution (IVR) on the selectivity of the experiment, and (iii) the full simulation of the two-dimensional 2D-IR-UV/Vis experiment. State-of-the-art multidimensional wavepacket propagation techniques will be employed, in particular the Multiconfiguration Time-Dependent Hartree (MCTDH) method.
The PhD student will actively interact with her/his colleagues of project A4.1 (orthogonal uncaging strategies based on the VIPER pulse sequence) and project A2.1 (synthesis of novel caging groups). At an international level, the project involves a collaboration with Prof. Fabrizio Santoro (University of Pisa).
Suggested literature:
- "Mixed IR/VIS two-dimensional spectroscopy: chemical exchange beyond the vibrational lifetime and sub-ensemble selective photochemistry",
L. J. G. W. van Wilderen, A. T. Messmer, J. Bredenbeck, Angew. Chem. Int. Ed. 2014, 126, 2705-2710. - "Using the MCTDH wavepacket propagation method to describe multi-mode nonadiabatic dynamics",
G. A. Worth, H.-D. Meyer, H. Köppel, L. S. Cederbaum, I. Burghardt, Int. Rev. Phys. Chem. 2008, 27, 569-606.
A1.3: Photochemistry of Porphyrin and Coumarin Systems
Photochemistry of Porphyrin and Coumarin Compounds
This PhD thesis addresses the complex photochemistry of uncaging of coumarin and porphyrin based molecular assemblies, employing electronic structure as well as molecular dynamics (MD) and quantum dynamical techniques.
The first focus area involves the characterization of novel coumarin-based dyads with enhanced two-photon absorption properties, in collaboration with the Heckel group (A2.2) and the Wachtveitl group (A3.2). The key role of intermolecular interactions will be studied, highlighting the role of competing energy and charge transfer pathways.
The second focus area concerns the photochemistry of free-base porphyrins and metal–porphyrins, notably in heme complexes. Both types of porphyrins are important building blocks in bioengineering and in the design of artificial light-harvesting materials. Despite the ubiquitous occurrence of porphyrins, key issues regarding their photochemistry are still open. In particular, the PhD student will investigate the ultrafast internal conversion mechanisms and their interplay with uncaging dynamics. In a collaboration with the Schlichting group (MPI for Medical Research, Heidelberg) and the Hummer group (MPI for Biophysics, Frankfurt), the PhD student will participate in the investigation of the photolysis, i.e., photoinduced uncaging, of CO from the prosthetic heme group of myoglobin. This state-of-the-art study focuses on time-resolved experiments undertaken at an X-ray free-electron laser, and gives unprecedented insight into the collective protein motions ("protein quake") triggered by CO photolysis.
A1.4: Multi-scale modeling of uncaging dynamics
The computational study and optimization of uncaging platforms involves the primary photoexcitation step, the photocleavage kinetics, and conformational changes triggered by uncaging. In this project, these three steps will be addressed in the context of a
novel intramolecular circularization strategy that was demonstrated for oligonucleotides by Patrick Seyfried (A2.4) during the first CLiC term. This new photo-tethering strategy is a particularly promising approach for DNA photocontrol since long oligonucleotide sequences can be caged in a controlled and reversible manner.
The PhD student will employ selected electronic structure calculations and hybrid quantum mechanics/molecular mechanics (QM/MM) approach, combined with molecular dynamics simulations. Furthermore, Markov State modeling techniques will be applied in collaboration with Prof. Bettina Keller (FU Berlin) to assess DNA conformational changes and destabilization on longer time scales. The project is closely connected to A2.7 as well as spectroscopic investigations by the A3 and A4 teams.
 A1.5: Simulation and optimization of two-photon and multi-quantum VIPER uncaging
A1.5: Simulation and optimization of two-photon and multi-quantum VIPER uncaging
Uncaging induced by VIPER (VIbrationally Promoted Electronic Resonance) excitation is a powerful approach that combines selectivity in the infrared (IR) domain with optical (UV/VIS) excitation. This approach was pioneered by the A4 spectroscopy team, in collaboration with our theory group (A1). As demonstrated during the first CLiC term by Daniela Kern-Michler (A4.1) and Carsten Neumann (A4.2) together with Jan von Cosel (A1.1), selectivity can even be achieved among different isotopologues. On the theory side, the first-principles investigation of vibrational-electronic (vibronic) couplings allows to make predictions of mode-specific VIPER efficiencies.
In the present PhD project, the potential of VIPER uncaging will be further explored by turning to two-photon VIPER activation, along with multi-quantum vibrational excitation, and vibrational energy transfer, in a teamwork effort with the spectroscopy group (A4.3 and A4.4). The PhD student will also interact with the synthesis group (A2.5 and A2.6), as well as our external collaboration partner Dr. Fabrizio Santoro (Pisa). Both known uncaging architectures and new platforms will be investigated, employing state-of-the-art electronic structure and quantum dynamical methods that are combined to understand the vibronic effects that drive VIPER spectroscopy. The PhD student will develop a predictive simulation platform that also includes the full simulation of the two-dimensional 2D-IR-UV/Vis experiment.



